
Philip Moriarty
This is a guest post by Philip Moriarty, Professor of Physics at the University of Nottingham
A few days ago, Raphael highlighted the kerfuffle that our paper, Critical assessment of the evidence for striped nanoparticles, has generated over at PubPeer and elsewhere on the internet. (This excellent post from Neuroskeptic is particularly worth reading – more on this below). At one point the intense interest in the paper and associated comments thread ‘broke’ PubPeer — the site had difficulty dealing with the traffic, leading to this alert:
This thread is generating unprecedented interest in PubPeer. Please bear with us as we deal with the traffic. https://t.co/lwWdMrDzAN
— PubPeer (@PubPeer) January 7, 2014
At the time of writing, there are seventy-eight comments on the paper, quite a few of which are rather technical and dig down into the minutiae of the many flaws in the striped nanoparticle ‘oeuvre’ of Francesco Stellacci and co-workers. It is, however, now getting very difficult to follow the thread over at PubPeer, partly because of the myriad comments labelled “Unregistered Submission” – it has been suggested that PubPeer consider modifying their comment labelling system – but mostly because of the rather circular nature of the arguments and the inability to incorporate figures/images directly into a comments thread to facilitate discussion and explanation. The ease of incorporating images, figures, and, indeed, video in a blog post means that a WordPress site such as Raphael’s is a rather more attractive proposition when making particular scientific/technical points about Stellacci et al.’s data acquisition/analysis protocols. That’s why the following discussion is posted here, rather than at PubPeer.
Unwarranted assumptions about unReg?
Julian Stirling, the lead author of the “Critical assessment…” paper, and I have spent a considerable amount of time and effort over the last week addressing the comments of one particular “Unregistered Submission” at PubPeer who, although categorically stating right from the off that (s)he was in no way connected with Stellacci and co-workers, nonetheless has remarkably in-depth knowledge of a number of key papers (and their associated supplementary information) from the Stellacci group.
It is important to note that although our critique of Stellacci et al.’s data has, to the best of our knowledge, attracted the greatest number of comments for any paper at PubPeer to date, this is not indicative of widespread debate about our criticism of the striped nanoparticle papers (which now number close to thirty). Instead, the majority of comments at PubPeer are very supportive of the arguments in our “Critical assessment…” paper. It is only a particular commenter, who does not wish to log into the PubPeer site and is therefore labelled “Unregistered Submission” every time they post (I’ll call them unReg from now on), that is challenging our critique.
We have dealt repeatedly, and forensically, with a series of comments from unReg over at PubPeer. However, although unReg has made a couple of extremely important admissions (which I’ll come to below), they continue to argue, on entirely unphysical grounds, that the stripes observed by Stellacci et al. in many cases are not the result of artefacts and improper data acquisition/analysis protocols.
unReg’s persistence in attempting to explain away artefacts could be due to a couple of things: (i) we are being subjected to a debating approach somewhat akin to the Gish gallop. (My sincere thanks to a colleague – not at Nottingham, nor, indeed, in the UK – who has been following the thread at PubPeer and suggested this to us by e-mail. Julian also recently raised it in a comment elsewhere at Raphael’s blog which is well worth reading); and/or (ii) our assumption throughout that unReg is familiar with the basic ideas and protocols of experimental science, at least at undergraduate level, may be wrong.
Because we have no idea of unReg’s scientific background – despite a couple of commenters at PubPeer explicitly asking unReg to clarify this point – we assumed that they had a reasonable understanding of basic aspects of experimental physics such as noise reduction, treatment of experimental uncertainties, accuracy vs precision etc… But Julian and I realised yesterday afternoon that perhaps the reason we and unReg keep ‘speaking past’ each other is because unReg may well not have a very strong or extensive background in experimental science. Their suggestion at one point in the PubPeer comments thread that “the absence of evidence is not evidence of absence” is a rather remarkable statement for an experimentalist to make. We therefore suspect that the central reason why unReg is not following our arguments is their lack of experience with, and absence of training in, basic experimental science.
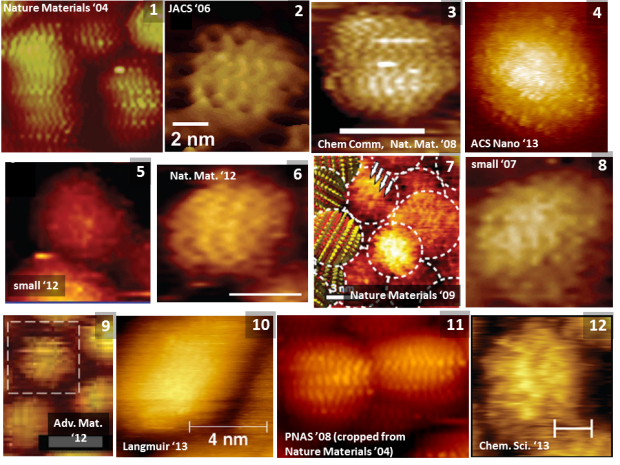
As such, I thought it might be a useful exercise – both for unReg and any students who might be following the debate – to adopt a slightly more tutorial approach in the discussion of the issues with the stripy nanoparticle data so as to complement the very technical discussion given in our paper and at PubPeer. Let’s start by looking at a selection of stripy nanoparticle images ‘through the ages’ (well, over the last decade or so).
The Evolution of Stripes: From feedback loop ringing to CSI image analysis protocols
The images labelled 1 – 12 below represent the majority of the types of striped nanoparticle image published to date. (I had hoped to put together a 4 x 4 or 4 x5 matrix of images but, due to image re-use throughout Stellacci et al.’s work, there aren’t enough separate papers to do that).
Putting the images side by side like this is very instructive. Note the distinct variation in the ‘visibility’ of the stripes. Stellacci and co-workers will claim that this is because the terminating ligands are not the same on every particle. That’s certainly one interpretation. Note, however, that images 1, 2, 4, and 11 each have the same type of octanethiol- mercaptopropionic acid (2:1) termination and that we have shown, through an analysis of the raw data, that images #1 and #11 result from a scanning tunnelling microscopy artefact known as feedback loop ringing (see p.73 of this scanning probe microscopy manual).
A key question which has been raised repeatedly (see, for example, Peer 7’s comment in this sub-thread) is just why Stellacci et al., or any other group (including those selected by Francesco Stellacci to independently verify his results), has not reproduced the type of exceptionally high contrast images of stripes seen in images #1,#2,#3, and #11 in any of the studies carried out in 2013. This question still hangs in the air at PubPeer…
Moreover, the inclusion of Image #5 above is not a mistake on my part – I’ll leave it to the reader to identify just where the stripes are supposed to lie in this image. Images #10 and #12 similarly represent a challenge for the eagle-eyed reader, while Image #4 warrants its own extended discussion below because it forms a cornerstone of unReg’s argument that the stripes are real. Far from supporting the stripes hypothesis, however, Stellacci et al’s own analysis of Image #4 contradicts their previous measurements and arguments (see “Fourier analysis or should we use a ruler instead?” below).
What is exceptionally important to note is that, as we show in considerable detail in “Critical assessment…”, a variety of artefacts and improper data acquisition/analysis protocols – and not just feedback loop ringing – are responsible for the variety of striped images seen above. For those with no experience in scanning probe microscopy, this may seem like a remarkable claim at first glance, particularly given that those striped nanoparticle images have led to over thirty papers in some of the most prestigious journals in nanoscience (and, more broadly, in science in general). However, we justify each of our claims in extensive detail in Stirling et al. The key effects are as follows:
– Feedback loop ringing (see, for example, Fig. 3 of “Critical assessment…”. Note that nanoparticles in that figure are entirely ligand-free).
– The “CSI” effect. We know from access to (some of) the raw data that a very common approach to STM imaging in the Stellacci group (up until ~ 2012) was to image very large areas with relatively low pixel densities and then rely on offline zooming into areas no more than a few tens of pixels across to “resolve” stripes. This ‘CSI’ approach to STM is unheard of in the scanning probe community because if we want to get higher resolution images, we simply reduce the scan area. The Stellacci et al. method can be used to generate stripes on entirely unfunctionalised particles, as shown here.
– Observer bias. The eye is remarkably adept at picking patterns out of uncorrelated noise. Fig. 9 in Stirling et al. demonstrates this effect for ‘striped’ nanoparticles. I have referred to this post from my erstwhile colleague Peter Coles repeatedly throughout the debate at PubPeer. I recommend that anyone involved in image interpretation read Coles’ post.
Fourier analysis or should we use a ruler instead?
I love Fourier analysis. Indeed, about the only ‘Eureka!’ moment I had as an undergraduate was when I realised that the Heisenberg uncertainty principle is nothing more than a Fourier transform. (Those readers who are unfamiliar with Fourier analysis and might like a brief overview could perhaps refer to this Sixty Symbols video, or, for much more (mathematical) detail, this set of notes I wrote for an undergraduate module a number of years ago).
In “Critical assessment…” we show, via a Fourier approach, that the measurements of stripe spacing in papers published by Stellacci et al in the period from 2006 to 2009 – and subsequently used to claim that the stripes do not arise from feedback loop ringing – are comprehensively incorrectly estimated. We are confident in our results here because of a clear peak in our Fourier space data (See Figures S1 and S2 of the paper).
Fabio Biscarini and co-workers, in collaboration with Stellacci et al, have attempted to use Fourier analysis to calculate the ‘periodicity’ of the nanoparticle stripes. They use Fourier transform of the raw images, averaged in the slow scan direction. No peak is visible in this Fourier space data, even when plotting on a logarithmic scale in an attempt to increase contrast/visibility. Instead, the Fourier space data just shows a decay with a couple of plateaus in it. They claim – erroneously, for reasons we cover below – that the corners of the second plateau and the continuing decay (called a “shoulder” by Biscarini et al.) indicates stripe spacing. To locate these shoulders they apply a fitting method.
We describe in detail in “Critical assessment…” that not only is the fitting strategy used to extract the spatial frequencies highly questionable – a seven free-parameter fit to selectively ‘edited’ data is always going to be somewhat lacking in credibility – but that the error bars on the spatial frequencies extracted are underestimated by a very large amount.
Moreover, Biscarini et al. claim the following in the conclusions of their paper:
“The analysis of STM images has shown that mixed-ligand NPs exhibit a spatially correlated architecture with a periodicity of ∼1 nm that is independent of the imaging conditions and can be reproduced in four different laboratories using three different STM microscopes. This PSD [power spectral density; i.e. the modulus squared of the Fourier transform] analysis also shows…”
Note that the clear, and entirely misleading, implication here is that use of the power spectral density (PSD – a way of representing the Fourier space data) analysis employed by Biscarini et al. can identify “spatially correlated architecture”. Fig. 10 of our “Critical assessment…” paper demonstrates that this is not at all the case: the shoulders can equally well arise from random speckling.
This unconventional approach to Fourier analysis is not even internally consistent with measurements of stripe spacings as identified by Stellacci and co-workers. Anyone can show this using a pen, a ruler, and a print-out of the images of stripes shown in Fig. 3 of Ong et al. It’s essential to note that Ong et al. claim that they measure a spacing of 1.2 nm between the ‘stripes’; this 1.2 nm figure is very important in terms of consistency with the data in earlier papers. Indeed, over at PubPeer, unReg uses it as a central argument of the case for stripes:
“… the extracted characteristic length from the respective fittings results in a characteristic length for the stripes of 1.22 +/- 0.08. This is close to the 1.06 +/-0.13 length for the stripes of the images in 2004 (Figure 3a in Biscarini et al.). Instead, for the homoligand particles, the number is much lower: 0.76 +/- 0.5 [(sic). unReg means ‘+/- 0.05’ here. The unit is nm] , as expected. So the characteristic lengths of the high resolution striped nanoparticles of 2013 and the low resolution striped nanoparticles of 2004 match within statistical error, ***which is strong evidence that the stripe features are real.***”
Notwithstanding the issue that the PSD analysis is entirely insensitive to the morphology of the ligands (i.e. it cannot distinguish between stripes and a random morphology), and can be abused to give a wide range of results, there’s a rather simpler and even more damaging inconsistency here.
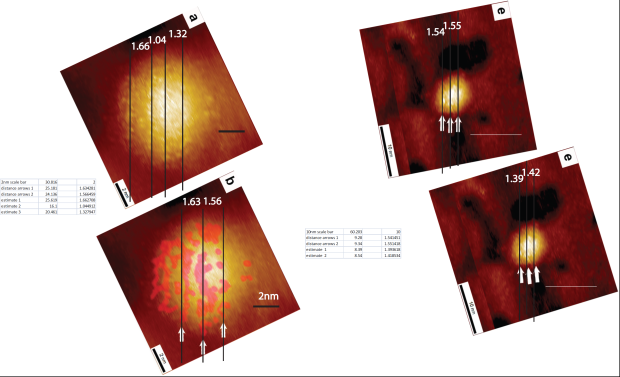
A number of researchers in the group here at Nottingham have repeated the ‘analysis’ in Ong et al. Take a look at the figure below. (Thanks to Adam Sweetman for putting this figure together). We have repeated the measurements of the stripe spacing for Fig. 3 of Ong et al. and we consistently find that, instead of a spacing of 1.2 nm, the separation of the ‘stripes’ using the arrows placed on the image by Ong et al. themselves has a mean value of 1.6 nm (± 0.1 nm). What is also interesting to note is that the placement of the arrows “to guide the eye” does not particularly agree with a placement based on the “centre of mass” of the features identified as stripes. In that case, the separation is far from regular.
We would ask that readers of Raphael’s blog – if you’ve got this far into this incredibly long post! – repeat the measurement to convince yourself that the quoted 1.2 nm value does not stand up to scrutiny.
So, not only does the PSD analysis carried out by Biscarini et al. not recover the real space value for the stripe spacing (leaving aside the question of just how those stripes were identified), but there is a significant difference between the stripe spacing claimed in the 2004 Nature Materials paper and that in the 2013 papers. Both of these points severely undermine the case for stripy nanoparticles. Moreover, the inability of Ong et al. to report the correct spacing for the stripes from simple measurements of their STM instruments raises significant questions about the reliability of the other data in their paper.
As the title of this post says, whither stripes?
Reducing noise pollution
A very common technique in experimental science to increase signal-to-noise (SNR) ratio is signal averaging. I have spent many long hours at synchrotron beamlines while we repeatedly scanned the same energy window watching as a peak gradually appeared from out of the noise. But averaging is of course not restricted to synchrotron spectroscopy – practically every area of science, including SPM, can benefit from the advantages of simply summing a signal over the course of time.
A particularly frustrating aspect of the discussion at PubPeer, however, has been unReg’s continued assertion that even though summing of consecutive images of the same area gives rise to completely smooth particles (see Fig. 5(k) of “Critical assessment…”), this does not mean that there is no signal from stripes present in the scans. This claim has puzzled not just Julian and myself, but a number of other commenters at PubPeer, including Peer 7:
“If a feature can not be reproduced in two successive equivalent experiments then the feature does not exist because the experiment is not reproducible. Otherwise how do you chose between two experiments with one showing the feature and the other not showing it? Which is the correct one ? Please explain to me.
Furthermore, if a too high noise is the cause of the lack of reproducibility than the signal to noise ratio is too low and once again the experiment has to be discarded and/or improved to increase this S/N. Repeating experiments is a good way to do this and if the signal does not come out of the noise when the number of experiment increases than it does not exist.
This is Experimental Science 101 and may (should) seem obvious to everyone here…”
I’ve put together a short video of a LabVIEW demo I wrote for my first year undergrad tutees to show how effective signal averaging can be. I thought it might help to clear up any misconceptions…
The Radon test
There is yet another problem, however, with the data from Ong et al. which we analysed in the previous section. This one is equally fundamental. While Ong et al. have drawn arrows to “guide the eye” to features they identify as stripes (and we’ve followed their guidelines when attempting to identify those ‘stripes’ ourselves), those stripes really do not stand up tall and proud like their counterparts ten years ago (compare images #1 and #4, or compare #4 and #11 in that montage above).
Julian and I have stressed to unReg a number of times that it is not enough to “eyeball” images and pull out what you think are patterns. Particularly when the images are as noisy as those in Stellacci et al’s recent papers, it is essential to try to adopt a more quantitiative, or at least less subjective approach. In principle, Fourier transforms should be able to help with this, but only if they are applied robustly. If spacings identified in real space (as measured using a pen and ruler on a printout of an image) don’t agree with the spacings measured by Fourier analysis – as for the data of Ong et al. discussed above – then this really should sound warning bells.
One method of improving objectivity in stripe detection is to use a Radon transform (which for reasons I won’t go into here – but Julian may well in a future post! – is closely related to the Fourier transform). Without swamping you in mathematical detail, the Radon transform is the projection of the intensity of an image along a radial line at a particular angular displacement. (It’s important in, for example, computerised tomography). In a nutshell, lines in an image will show up as peaks in the Radon transform.
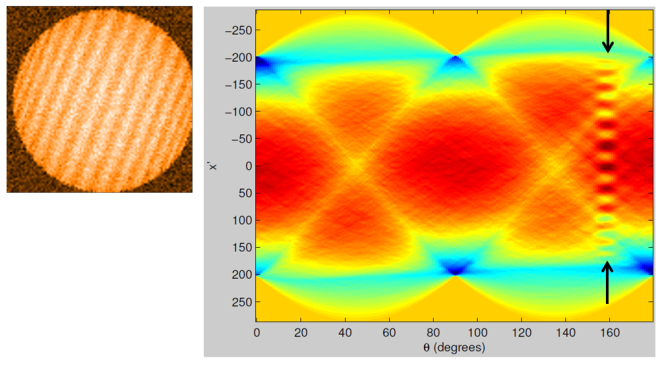
So what does it look like in practice, and when applied to stripy nanoparticle images? (All of the analysis and coding associated with the discussion below are courtesy of Julian yet again). Well, let’s start with a simulated stripy nanoparticle image where the stripes are clearly visible – that’s shown on the left below and its Radon transform is on the right.
Note the series of peaks appearing at an angle of ~ 160°. This corresponds to the angular orientation of the stripes. The Radon transform does a good job of detecting the presence of stripes and, moreover, objectively yields the angular orientation of the stripes.
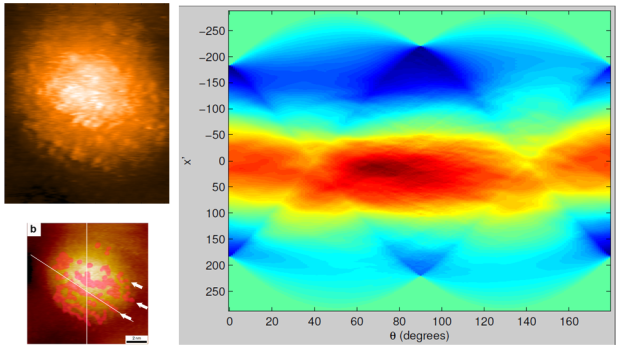
What happens when we feed the purportedly striped image from Ong et al. (i.e. Image #4 in the montage) into the Radon transform? The data are below. Note the absence of any peaks at angles anywhere near the vicinity of the angular orientation which Ong et al. assigned to the stripes (i.e. ~ 60°; see image on lower left below)…
Hyperempiricism
If anyone’s still left reading out there at this point, I’d like to close this exceptionally lengthy post by quoting from Neuroskeptic’s fascinating and extremely important “Science is Interpretation” piece over at the Discover magazine blogging site:
“The idea that new science requires new data might be called hyperempiricism. This is a popular stance among journal editors (perhaps because it makes copyright disputes less likely). Hyperempiricism also appeals to scientists when their work is being critiqued; it allows them to say to critics, “go away until you get some data of your own”, even when the dispute is not about the data, but about how it should be interpreted.”
Meanwhile, back at PubPeer, unReg has suggested that we should “… go back to the lab and do more work”.
I have also posted this comment over at PubPeer to directly address each of unReg’s ‘criticisms’ of our paper: https://pubpeer.com/publications/B02C5ED24DB280ABD0FCC59B872D04/comments/5593
LikeLike
Killer post! I could follow it all.
Minor thing: Interesting to look at the page 73 of SPM manual warning of feedback ringing ‘especially on high slope areas’. And then we are looking at nanoparticles (high slope).
The Radon transform is quite nice. Really, when looking at images like 4, they are on shaking ground drawing in lines and saying one has something versus just chance blotches lining up a little. Adding onto that, one presumably has the issue of cherrypicking which particles to look at from the overall image.
LikeLike
The Radon transform approach was entirely Julian’s idea – I can’t take any credit for that. It’s pretty neat, isn’t it? My final year undergraduate project – back in 1990 – was on computerised tomography so in the dim distant past I had encountered the Radon transform in the context of backprojection and CT.
LikeLike
One curiosity (not sure how this fits in the drama): why do the stripes in 1, 3, and 11 (the classic stripe images) RIPPLE so much? Surely that would not be expected just from the geometry? Did the authors address this in their paper? I saw how the ripples were similar in the simulated (and tested on bare gold) feedback ringing. What makes it not just have stripes, but ripply stripes?
LikeLike
That’s a very perceptive observation! The reason they ripple is, of course, because they arise from feedback loop ringing. To be fair to Stellacci et al, they did call the features “ripples” and attempted, on the basis of an analysis which we show in “Critical assessment…” is thoroughly flawed, to separate ripples from noise. See Fig. 3 of our paper for examples of those types of ripples created by high feedback loop gains on entirely ligand-free particles.
LikeLike
Oh…it seems really telling that they have 30 papers but so few actual images (and then the scandal of the uncited reuse of data in some papers).
It really seems like they have found a few places where they thought they saw this…then got so attached to the hypothesis…and built a house of cards on top of it (other ligands, drug delivery, etc.). But at the end of the day, they are just selling themselves based off of that mistake from 2004 (which itself doesn’t always show up all over the images…the zoom thing) and was pointed out to them in 2005.
I have to wonder how his students feel, postdocs feel. Wonder if students (last couple years) have started to steer clear of the guy when it comes to picking an advisor…I bet he is not getting the pick of the litter.
LikeLike
Wow, great post. How could it be clearer than what you’ve shown in the figure?
LikeLike
Thanks, nanoymous – I’m delighted that you liked it. I suspect, however, that we have not heard the last from “unReg”…
In any case, I’m putting the stripy saga to one side now until we get the referees’ reports from PlosONE.
Best wishes,
Philip
LikeLike
Good post. From my non-expert’s point of view, there is just one fundamental question here: why have the clearly visible stripes seen on earlier images disappeared on more recent ones?
To me the question of whether one can detect subtle stripes in the recent images using fancy mathematics is a moot point – the very fact that one needs to resort to such measures is a testament to the fact that the original, visible-with-the-naked-eye stripes have vanished.
LikeLike
That is a question which experts have asked to. The problem here is Stellacci et al. and “the defender” on PubPeer insist that the stripes ARE clear on the new images. It just becomes surreal. On PubPeer I was told “Well, then Stirling will have to show evidence that they do not look like stripes, because they very much do” talking about the very image I Radon transformed above. I entirely agree that the fancy mathematics is not needed, but it was used because the debate about these images had essentially descended to:
“But those aren’t even stripy”
“Are too!”
“Are not”
“Are too!”
The maths was introduced out of desperation for the rather surreal purpose of arguing to scientists that these images don’t look stripy.
LikeLike
thanks @Neuro-Skeptic. You are absolutely right that there should be no need, but, in addition to the surreal discussion at PubPeer (as highlighted by Julian), there are the surreal 2013 articles published in highly respected journals (ACS Nano and Langmuir), which, using “such measures” claim that the vanished stripes are there… even if you can’t see them.
Those articles were approved by editors. They went through the peer review process, so, presumably, they were approved by independent expert referees. Somehow, both referees and editors failed to ask the one fundamental question you highlight.
The ACS Nano ms was published alongside a particularly timely editorial (without direct reference to the said article to be clear) which criticized scrutiny of published articles on blogs (http://blog.chembark.com/2013/10/23/response-to-acs-nano-editorial-on-reporting-misconduct/).
LikeLike
Perhaps the solution is a double blind test.
Get some neutral observers who have never seen these images before. They wouldn’t even need to know STM.
Show them the images and ask them (in this order) a) what do you see? b) do you see stripes? c) assuming that there are stripes here, what is their orientation?
I suspect that most people would see stripes in the early images, but not in the later ones; or if they did ‘see’ them, they would not agree on the orientation. And also I suspect that people would not distinguish between later “stripey” and “control” images.
LikeLike
@Neuro_Skeptic
That is indeed possible. The problem though is that humans are brilliant at seeing patterns where there are none. Phil often links to this post: http://telescoper.wordpress.com/2009/04/04/points-and-poisson-davril/ here the one that appears to most “neutral observers” to be random was generated with weighting towards spacing. The one that has the clear clustering is truly random. As such fancy maths is more reliable that humans for objectively recognising randomness verses clustering. Our brains are so obsessed with patterns we find them when they are not there.
LikeLike
Nice post. This subject should be thoroughly dealt with by now! It’s interesting looking back at that “old” document “A Practical Guide to SPM” There’s some great stuff in there. Particularly relevant is page 75, on which we see some great advice:
“Whenever you are suspicious that an image may contain artifacts, follow these steps:
1. Repeat the scan to ensure that it looks the same.
2. Change the scan direction and take a new image.
3. Change the scan size and take an image to ensure that the features scale
properly.
4. Rotate the sample and take an image to identify tip imaging (see Figure 4-2).
5. Change the scan speed and take another image (especially if you see
suspicious periodic or quasi-periodic features).”
The relevance of this is obvious – if these steps has been followed in the first place we would not still be here discussing this.
LikeLike
Excellent post, reminded me of the good old days when I was STMing myself!
One minor thing: I think the link to the paper by Fabio Biscarini is not correct (it’s the other paper of 2013).
I really hope all of you continue your efforts to make sure the scientific community will finally realize that there ar NO striped nanoparticles. I remember discussing these papers with other SPM-people back in the days and none of them had any doubts that there were obvious errors in these papers. However, nobody really bother to correct this because everybody just thought “these mistakes are so obvious, nobody will take these papers seriously”. Well, looks like we were wrong… So please keep up the good work!
Personally I think that one of the reasons that all of this happened is because many users underestimate the difficulty / complexity of SPM becaue it’s a very ‘visual’ technique. When images are aquired properly, it is often easy to show also to non-SPM-experts what you can see in these images. I think this a speciality of SPM techniques (maybe it’s similar for SEM/TEM but I’m not an expert in that field) because for many other – comparably complex – exmperimental techniques it is not so straightforward to explain the results to a non-specialist because of the lack of visual material. It can then happen that non-specialists get the impression that interpreting SPM-data is ‘easy’ (which it can be sometimes if you have made sure that there are no artifacts during data acquisition) while in reality it takes a couple of years to be able to correctly ‘read’ SPM-images. Because of this seemingly trivial nature of SPM, some new users believe that it is a ‘plug-and-play’ scientific technique (there are some analytical instruments which you can order from a company, get them installed in your lab, do a 2-day workshop and they are ready to use, I call that ‘plug-and-play’ science).
In view of the above I can also understand the sometimes stern tone (I’m obviously not a native English speaker so I don’t know if this is an appropriate word here) in the replies by Moriarty & Co in the comments. I can see how frustrating it is if people will not see the complexity of a technique and – even though they know very little about the details of SPM – tell you that you are wrong.
LikeLike
Thanks for your encouraging words and for pointing the wrong link (now corrected).
LikeLike
Moriarty,
Thank you very much for the detailed responses to most of my points on the blog of Levy, and for the summary here on PubPeer. Much appreciated.
However, your answers are far from satisfactory and do not address the issues I raised. Actually, they lead to even more concerns about the arguments in your paper. The only conclusion that one can make is that the thesis and conclusions in the paper don’t hold water. But let me go directly to the technical points; will start with the more important ones (please not that a few of my points below are a reply to more than one of your numbered points in your post of yesterday):
1. The Radon transform. You wrote “if we try to objectively find evidence for stripes in the data of Ong et al. via a Radon transform, we draw a blank”. Well, no. Actually, your Radon transform of the striped image from Ong et al. (Figure 7b in your paper) shows the presence of the stripes. Let’s go step by step:
The Radon transform of your simulated striped nanoparticle in https://raphazlab.files.wordpress.com/2014/01/radon-1.png nicely shows all the stripes at about 160 deg (the direction perpendicular to that aligned with the stripes) as a periodic line of alternating light green / dark red marks, as one would expect from a line integral along a straight line (that is, a Radon transform). Now, the real STM image is of course much less defined than your clean simulated image, so one would expect a much less obvious pattern. Another difference from your simulated image and the STM image is that you drew striped bands instead of lines of dots (the individual ligands in the high-resolution STM). A much more appropriate comparison would have been a simulated image of parallel lines of dots (instead of parallel bands), where all the dots in a line do not align with the dots in consecutive lines (as it occurs with the real STM images).
In fact, because the dots from different stripes do not necessarily align, it is not surprising that the line integral at 60 deg over the whole image is absent of features (bright dots and darker regions between dots roughly cancel out). However, it is clear from the STM image that the spacing of the dots within a single line of dots is relatively constant. So the line integral along the direction of the line of dots with consistent spacing should leave a fingerprint. Indeed, the white line you have drawn on top of a line of dots on panel b in https://raphazlab.files.wordpress.com/2014/01/radon-2.png is at 140 deg. And what do you see at 140 deg on the Radon transform? A few alternating yellow / green marks. They are week, as expected, but can be seen clearly. This is the signature of the lines of dots, that is, stripes, in the STM image.
* So your Radon transform analysis shows that there are striped features in Figure 7b in your paper. *
2. Stripe spacing in the high-resolution images of Ong et al. You wrote “A cornerstone of your argument is therefore shown to be entirely incorrect: the spacings that you get from the PSD analysis do not agree with the spacings measured from the Ong et al. images shown in Fig. 3.” This is wrong. In the paper, Ong et al. state that THE AVERAGE distance between stripes is about 1.2, but that the spacing of the stripes in the high-resolution images ranges from ~0.7 to ~1.5. You used a ruler to measure a few distances, but if you look at the average line profiles in Figure S10b of Ong et al., a much more precise approach, you will see that the average distance between stripes is close to 1.2.
* So the spacing of the stripes from real space analysis (Figure S10b in Ong et al.) and reciprocal space analysis (Figure S10a in Ong et al.) match. *
3. The fitting of the PSDs. I have to say that your Figure S5 is hugely misleading. You haven’t fitted the magenta and red curved properly within the shoulder region of the PSD, that is, you have not set a sufficiently low tolerance for the root mean square deviation of the fit, so of course you obtain a large variability in the parameters. You can’t calculate the spread of the fitted wave vector by using badly fitted curves. If you do the fitting properly, you should get the values provided in the paper: 1.06 +/- 0.13.
* Your Figure S5 is an example of substandard regression analysis. It invalidates your argument that reliable stripe spacings can’t be extracted from the PSDs*
4. Referring to the 2006 work published in JACS, you wrote “the ringing in the current image can be identified as strong peaks in the PSD (Figure S2 and S3) which is then used to show that the eye-balling of the “real” stripes in that paper was flawed (Fig 4) as they are within frequency range of the strong oscillations (ringing) in the current image.”
However, in your Figure 4 you plot the spatial frequency range for the noise from the tunnel current images corresponding to Figure 3 of the 2006 work (JACS). But you do not say in your paper that the images that you analyze contain both nanoparticles and flat regions (the gold foil substrate), and that the substrate largely dominates (in percentage of image area). So when you analyze the noise, you are mostly analyzing the noise corresponding to the substrate. Stellacci mentions in the paper that when imaging the substrate, the features are tip-speed dependent, which means that the ripples arise from noise, but when imaging the area where the nanoparticles sit, the features are NOT tip-speed dependent, which rules out noise artifacts. So Stellacci shows that stripe features arising from noise can be distinguished from those that are genuine by looking at how the spacing of the stripes changes with tip speed (the spacing doesn’t change for the ripples on the nanoparticles). But because the substrate in the images you analyze dominates, in your Figure 4 you are showing the frequency range of the noise corresponding to the substrate (in fact, the grey area and the red dots correlate and decrease in value with tip speed; this is not the case for the green diamonds and blue triangles, which are largely tip speed independent). Then you assume that this noise would be the same for the nanoparticles. This is wrong. The grey area in your Figure 4 is not the relevant noise for the green diamonds and blue triangles. You should extract the power spectra and relevant spatial frequencies for the nanoparticle regions only (not for the whole image!) to obtain the noise range for the nanoparticles and compare it with the spatial frequency for the nanoparticles (the green diamonds and blue triangles). To sum up this point, you are comparing apples (the noise range for the spatial features of the substrate) with pears (the spatial features of the nanoparticles).
Continuing with this point, there is another misrepresentation in your paper. You wrote “Jackson et al. 2006 [24] also state that the gold foil substrates used in the work have curvature comparable to that of the nanoparticle core. This begs the question as to just how some areas were objectively defined as the surface, and thus exhibited feedback noise, while others were defined as nanoparticles with molecular resolution”. But this sentence quoted from the JACS paper refers to Figure 1, not to Figure 3, which is the one you analyzed. Also, you ‘forgot’ to mention that the curved gold foil substrates do not show any features (just look at the circled area in Figure 1a of the JACS paper), which is not what happens to Figure 3a, where there are clear ripples on the substrates. So you again misrepresent facts to prove your points.
* So the comparison of error ranges in your Figure 4 is misleading, and therefore you have no support for your claim that the stripes can’t be distinguished from noise in the 2006 images *
5. Stellacci and co-authors have shown in Biscarini et al. that the images in 2004 (but for one outlier) are not tainted by feedback look ringing (and, as I showed above, your claims for the invalidity of the PSD analyses are based on poor use of regression analyses and statistics). You have only analyzed one image from 2004 for feedback-loop ringing (which may be such outlier). Therefore, you can only claim that the stripes in ONE 2004 image (not all of them) are likely to be arising from artifacts.
* So your claim that all of the 2004 work on striped nanoparticles is only a result of artifacts is unsubstantiated *
6. The low resolution 2004 images have been reproduced in a few papers, including refs. 24 and 47 (you claim that the striped nanoparticles in refs. 24 and 47 suffer from noise and observer bias problems, but as I detailed in the various criticisms above these arguments have many holes and do not stand up to scrutiny. In addition, the Biscarini et al. PSD analysis shows that the features in the high resolution images of 2013 and the low resolution images of 2004 are consistent, which is a strong argument for the reproducibility of the striped domains. Also, Stellacci did share the samples with the recognized STM experts Renner and De Feyter. You can find the comparison of images taken at the different labs in Fig. 1 of Biscarini et al.
* So your claim that the 2004 images have not been reproduced is just wrong *
7. You stated that you can’t answer my question of a few days ago, “Can you show me three aligned linear clusters of dots in a Poisson distribution of them? Can you point at closely aligned lines of dots in this image with random dots (https://telescoper.files.wordpress.com/2009/04/pointa.jpg?w=450&h=393)?”, because the areal number density of the Poisson point set has to match that of the ligand features for any compassion to make sense. Well, then you just have to rescale (compress or stretch) the Poisson point set image so as to match the areal density of the ligand features. It’s simple! In any case, you won’t be able to find closely aligned lines of dots systematically in a Poisson distribution of dots. You won’t.
* So your claim that the stripes can arise from Poisson noise is unfounded *
8. Your tutorial video on basic signal averaging shows clearly why you still don’t understand the logical blunder that you have committed with Figure 5 in the paper. When you do summation of 5 low-resolution images where the stripes cannot really be superposed (look at panels c and f in Figure 4 of Yu and Stellacci; after the appropriate image rotation the stripes can’t be properly superposed), you are NOT summing the same signal over and over as you do in your tutorial. To put it in a simplistic way, you are summing signal + noise + noise + noise + noise + … instead of signal + noise + signal + noise + signal + noise + … . In the latter, the noise cancels out with a large number of summations, as you show in the video. In the former, the signal necessarily gets weaker and disappears.
* So in low-resolution images where stripes do not appear consistently on the same place on the same nanoparticles, the summation approach makes the stripes weaker, not stronger. Your Figure 5 in the paper is therefore misleading. *
In conclusion, there are too many issues with the analyses in the paper. Your criticisms of the PSD analysis in Biscarini et al. are flawed, your analysis of the spatial frequency range for the stripes in the 2006 images is inappropriate, your assertions that stripes can arise from pure noise and that the 2004 images have not been reproduced are wrong, and your overall claim that “we CONCLUSIVELY show that ALL of the STM evidence for striped nanoparticles published to date can instead be explained by a combination of well-known instrumental artifacts, strong observer bias, and/or improper data acquisition/analysis protocols” (from the abstract of the paper) is, to say the least, wishful thinking. Moreover, Your Radon transform analysis actually shows the signature of stripes!
You should then go back to the lab and do more work. You have to redo Figure S5 properly, analyze quantitatively for observer bias in Figure 7b and the other high-resolution images in Ong et al., correct for the wrong error spread (grey area) in Figure 4 when comparing to the green diamonds and blue triangles, analyze for feedback-loop artifacts in a few more images, correct or eliminate Figure 5 as it is based on an inappropriate summation procedure, and rewrite the paper to remove unsupported statements. And if you do these things well, you will inevitably end up having no major criticisms of the evidence for stripes.
LikeLike
I have responded back at PubPeer.
Note also this comment over at PubPeer
Moriarty,
Thanks for the details (at PubPeer).
So you disregard the yellow / green marks at 140 deg in your Radon transform of the STM image of the high-resolution nanoparticle in Figure 3a of Ong et al., claiming that they arise from noise; that is, you claim that the Radon transform is doing an excellent job at pulling out noise in the image in the 140 deg direction. Well, once more you are wrong. I have done a few Radon transforms for you and put them together as an image here:
The image is self-explanatory, but I have added a few comments for you. Clearly, there is structure in the transform at 140 deg, and it corresponds to the stripes.
As for your comment on the average stripe width, your use of the ruler is far from precise, because you obtain significantly different numbers for exactly the same image! (for Figure 3a in Ong et al.: 1.66; 1.04; 1.32; and Figure 3b: 1.63; 1.56). So you did not do it properly; which shows that in these high-resolution images it is difficult to choose where to draw the lines over the striped features. Also, the width of the stripes is not reliable at the edges of the particle, where there is likely to be image distortion. This is why I said that a much better approach is to use the average line profiles in Figure S10b of Ong et al. In this case you get values really close to 1.2, within a small error bar.
To sum up, this wrong analyses of yours (Radon transform and careless measurements of stripe widths on the images) again highlights the way you arrive at your conclusions: lack of proper understanding of statistics, careless measurements, rushed interpretations, improper comparisons, hand picking of published data and inconsistent use of arguments across datasets.
LikeLike
This is hilarious to see unreg criticizing the crits. IT just totally goes without saying that the burden of proof is on the stripers. That they ran with feedback ringing in 2004. That they published 30 papers on a house of cards. and that they STILL don’t have conclusive proof of the stripes. Oh…if you squint and draw lines, you might have stripes. What junk. They had feedback ringing in 2004. When the resolution got good, the stripes disappeared. QED. Stop trying to squint into nothing, unreg.
LikeLike
Unreg,
It is nice to see that you’ve put up some figures and done some work which demonstrates to me that you are perhaps being sincere (though I must state my complete befuddlement at many of your statements about signal and noise, they do not seem credible to me).
Your final stripe lattice looks an awful lot like noise, would you mind adding a few lattices (say 10) that are generated completely randomly (and the associated radon transforms) to suggest how likely one is to see very weak (by your own admission) features in the Radon transform by chance?
LikeLike
Unreg,
It is nice to see that you’ve put up some figures and done some work which demonstrates to me that you are perhaps being sincere (though I must state my complete befuddlement at many of your statements about signal and noise, they do not seem credible to me).
Your final stripe lattice looks an awful lot like noise, would you mind adding a few lattices (say 10) that are generated completely randomly (and the associated radon transforms) to suggest how likely one is to see very weak (by your own admission) features in the Radon transform by chance?
LikeLike
Nanonymous,
I know I said I would stay out of the discussion. As you have mentioned the Radon transforms of UnReg I will just make a couple of little points on his Radon transforms.
First the reason that (s)he sees the features at a different angle from what we have suggested comes simply from setting up the Radon transform differently. Our is clearly set to integrate over lines rotated by theta anti-clockwise from the y-axis (you can see this by the stripe directions in our clearly striped simulation image). It is worth nothing that not only has UnReg set his/her transform up differently which is misleading, they have also not provided any axes which is unfortunate.
What is even more surprising is the way the transforms have been applied. Normally one takes a raw image, grey-scales it, subtracts the mean background (unless the background is zeros!), then applies the transform. The mean value subtraction is necessary due to the integration over angled lines in the image which will necessarily be of different lengths, this lack of subtraction is what gives rise to the strong XX shape dominating unRegs’ transforms. If the black and white ones had been inverted the clarity would have been a lot better as black is zero (or [0,0,0]) so the integration would have not given these crosses.
Even more surprising the images seem to have just been opened, in which ever program was used, as true-colour images and each channel seems to have been transformed separately then recombined to a bizarre true colour radon transform. This is why his transforms are so hard to read, and why they are the same colour as the image that produced them. An STM image is topography map, so 3 channel colour data is meaningless for any numerical calculations. One can take the intensity of each pixel to return to single channel data, take the transform of that (after appropriate subtraction), and then one would normally plot the radon transform to the full range of what ever colourmap one is using. This way they lower contrast white and grey dots would have lost no clarity as the signal to noise ratio has not changed!
The reason I critique the transforms is to make clear that unReg has just opened images, applied the transform quickly without learning how to correctly apply it. This generates very poor results with almost no contrast except the XX background (a little better on the one with the red overlay as the red the red comes through in the transform), and yet (s)he is happy state his/her conclusion on this very weak data, without a control image to compare against. When I was showing I can recognise stripes I also transformed a stripy image. If he is saying (s)he can see almost aligned dots are not noise, why not also transform Poisson distributed dots?
LikeLike
Yes completely agreed Julian.
Unreg, would love to see a few plots taking taking Julian’s advice into account (greyscale, mean subtraction, transformation of a stripy image etc.). Please also do post your snippet of code for generating the random distributions , looking forward to seeing your images.
LikeLike
Just wanted to say (since I slagged him pretty hard earlier) that I appreciate Unreg making the radon plots. It shows more than trolling, kudos.
That said, still seems like the guy is trying to justify a past mistake. Squinting into to the dot patterns for lines on something that had 30+ papers built on it, is not good news. Now we seem to have very varying stripe sizes, etc. Whole thing seems like trying to justify a signal out of noise and the burden of proof on the stripers and not shown.
The justification for keeping a view that stripes exist is that they are “really hard to resolve”. But then we have all this tissue paper evidence thrown together. Poor SEM, questionable statistics, poor TEM, NMR that does not show what is claimed, etc.
LikeLike
Stirling,
I did not bother to make the original images grayscale and subtract the mean background because it does not make much of a difference; the patterns can be appreciated anyway. Also, it is clear that an image with pure random noise will not show any defined features.
Still, I just grayscaled the images, subtracted the mean background, and performed the Radon transform again for the arrangements of dots, and for the same high resolution STM image. I have also added two random noise images and their Radon transforms as controls:
http://postimg.org/image/4w67w3y71
It is obvious that you and Moriarty are wrong when stating that the stripe features in Figure 7b of your paper arise from noise or observer bias.
LikeLike
Those middle images are incredibly faint. Yeah, there’s something on the radon. But those stripy smears could just be chance. Certainly like to see a sampling of more of the particles. FS well known to offline zoom and cherrypick parts of the image. And for that matter, how come the stripes look so much worse when the resolution is better? that 2004 image MUST be feedback ringing. Given that, I’m suspicious of superfaint random smears as “confirming” the original. Think it more likely cherrypicking and oversquinting. And pretty obvious why FS doesn’t want to share the nanoparticles for imaging (pretty cowardly).
LikeLike
Thank you unreg for considering julians requests. Could you please adjust things so that they are all comparable? The noise pictures arent noisy distributions of the small circles (with the same number of small circles). As it stands I can’t compare, the two types of images are categorically different. We need random distributions that correspond to you lattice-> noisy series (small black circles as glyphs). Also can add the 2004the stripy image?
LikeLike
Nanonymous,
Have done what you asked for: http://postimg.org/image/gh8pr3ect
As you will see, the Radon transforms capture the signatures of aligned dots or ripples as defined patterns at the appropriate angle(s), while random patterns show undefined, pretty homogeneous patterns across all angles.
LikeLike
Thank you Unreg,
My apologies for asking just a few last additions:
Can you also add the images of the unfunctionalized surfaces from Julian’s paper (top left in Figure 3)?
As well as the Langmuir ’13 image from Phillip’s post?
Again I apologize for the multiple requests and appreciate your accomodation.
FWIW I am having a hard time interpreting the Radon transforms so far. Failry clear on the first three, we see alternating low values (dark) and high values (the brightest in the image, clear if one squints).
On the 4th row (form Ong S9b) so see a sort of “zig zag” shape in the light blue oval. If I look all the way to the left I see a similar sort of “ziag zag” as well.
On the 5th row that is higher contrast, I can see the “bright-dark-bright-dark-bright-dark-bright” pattern a bit better.
One the 6th row I can see a faint “bright-dark-bright” pattern in approximately aligned with oval in the rows above.
On the 7th row there is a clear “bright-dark-bright” pattern perhaps just left of the oval position in previous images.
The second pair of images on the 8th row, there is a dark-bright-dark-bright pattern (again close to 140 deg…squint and you can see the two vertically aligned bright spots).
The positive controls are (2004 images) show “weaves” in the blue oval…the high contrast image in row 5 doesn’t seem to show this. What do you attribute the difference to?
Tough to cross compare since the images all have very different overall intensities. I hope the additional images might provide additional controls. I will happily respond in more detail if you could kindly accomdate my request at the top of the post, I hope you agree that those images are important controls.
LikeLike
I think we’ve moved from squinting at very poor stripes in images to squinting at small traces in radon. The issue is a lack of clear evidence. The two radons with the blue circles don’t show me a clear indication. I wonder if shown without the circles to clue the eye, if anyone would even get where there traces in the radon were. Actually even looking at them with the blue circles, not getting much jumping out at me.
I also remain concerned about the picking of individual particles given (as representative of the whole) given how utterly faint and arguable the stripes are. Really…the nanoparticles need to be shared.
LikeLike
Nanonymous,
As requested, I have updated the image of the Radon transforms with the unfunctionalized surface of Stirling et al., and Figures 2a and 2b of Ong et al. (Langmuir 2013):
http://postimg.org/image/aq2vku0rp
You will see that the transform of the unfunctionalized image doesn’t show any feature, as one expects, and that the transform of the striped nanoparticle of Ong et al. shows, when the contrast is high, marks at 140 deg, that is, the signature of the aligned dots.
In response to your comments above, of course there are bright and dark patterns in the images of random dots, one expects that. But the patterns are roughly homogeneous for the whole angle interval.
Signatures of alignment should appear as brighter features at specific angles (140 and 50 deg for the 2013 STM images of striped nanoparticles, and 90 deg for the 2004 STM images of striped nanoparticles). By the way, the patterns in the Radon transforms of course vary depending on where they are for round dots (neat examples), for blobs with undefined and variable shape (2013 STM images) or for ripples (2004 STM images).
LikeLike
Thank you Unreg for posting the additional images.
I agree that I can’t see much at all in Radon of the unfunctionalized surface.
I think it is very difficult for me to accept that looking at Radon transforms is conclusive in this case (at least how they’ve been done in the images that you have posted), or even informative past the obvious control images.
“You will see that the transform of the unfunctionalized image doesn’t show any feature, as one expects, and that the transform of the striped nanoparticle of Ong et al. shows, when the contrast is high, marks at 140 deg, that is, the signature of the aligned dots.”
I think this is misleading, the image(row 12 first column) that you describe as “high contrast” isn’t merely a high contrast image. You’ve clearly plotted surface glyphs (how? manually?) as perfect white circles. The contrast transform is rather harsh, the circles are all the same white but the the bottom of the image is clearly lighter than the top of the image.
I don’t think the Radon transform in the last row is materially different that the “noise” randon transform (row 8, 4th column).
I appreciate all the work you’ve done in getting these images up, but I must say that interpretation is really subjective at this point.
My understanding (from pubpeer) is that you believe the 2004 images (row 9) are correctly acquired (even though I believe you agree that there is feedback loop ringing in the 2004 image, do you think that is not material to the image quality?). They are clearly very different (the contrast is very clear, and not very clear in other images in row 4 & 11). What do you attribute this difference to?
LikeLike
Nanonymous,
Of course the Radon images are by themselves no ‘proof’ of the existence of stripes. The reason I did this exercise was to show Stirling and Moriarty that their claims about the stripes on the 2013 nanoparticles arising from observer bias or from noise are unfounded. Also, in most cases one does not really need the Radon images to see striped features on the high resolution STM images; but the Radon transforms give an objective measure of alignment and thus we avoid falling prey of our brains’ tendency to see patterns where they may be only noise. In the case of the high-resolution images of Stellacci, the Radon images add to the various evidences that there is alignment of dots, and therefore, stripe features.
Row 12, first column corresponds to Figure 2b of Ong et al, the only difference is that the circles are placed on a black background so as to enhance the contrast. If you see that the bottom half part of the transform is lighter is simply because the dots are not symmetrically distributed overall (there are more on the bottom half and on the left half). This is not the case for the rest of the images.
As I mentioned in previous comments, Radon transforms capture the signatures of aligned dots and stripes (which are aligned blobs or ripples, depending on whether they were taken with high or low resolution STM) as defined patterns at the appropriate angle(s), while random patterns show undefined, pretty homogeneous patterns across all angles. For example, if you look at the transform in the last row, the marks at 140 deg (inside the overlaid blue circle) are brighter than the immediate environment. You will also see in the same image similar patterns (in particular at 50-80 deg) as those in the transform in row 3. Instead, the images with randomly positioned dots don’t show any features that are clearly brighter and defined.
The transform of the 2004 image shows a clear bright-and-dark pattern because these are stripes (not aligned dots) that are clearly well aligned and with high contrast. Instead, the high resolution images correspond to individual ligands, the imaging of which is ‘effectively’ subject to more noise than the imaging of low-resolution contours (pre-2013 STM). This can be appreciated in the first three rows of transforms. As the local disorder increases, the features in the transform increasingly fade away. Also, a set of aligned dots has less ‘brightness’ than a solid stripe; this is why the contrast is worse for the high resolution 2013 images.
LikeLike
Thank you for you response Unreg.
I don’t mean to unilaterally change the path of the thread, but it seems like you do believe that the images acquired in 2004 are valid (in spite of the feedback) and are reproducible. If that is the case, all of the arguments from the Philip and the other members of the group essentially fall apart (by Phillip’s own admission, see 1k GBP bet).
In that case, we can avoid these sorts of debates around subjective features which (by your own admission, as I understand) are very close to the noise level.
Wouldn’t you agree that if Phillip re-imaged a batch of particles sent from Stellacci (and control imaging from a group like Renner) would essentially settle the argument? If the 2004 images could be reproduced using correct imaging, the stripes clearly would be reaffirmed and Phillip and his group simply would not have a whole lot of credibility after this outcome. If they could not be reproduced, that would seriously affect the validity of the 2004 paper and make the case against evidence for stripes very clear. If they could not be reproduced and the evidence for their existence rested on subjective interpretations of image features very close to the noise level, I think the outcome of the debate would be easily predicted.
Given the evidence of an ex-graduate student’s testimony (with respect to the particles as well as the referee reviews), the odd appearance of a Nature Materials editor on this site and the mysterious absence of a particle sample (forget about the scientific arguments, for the moment); I would hope that you would agree that there is good reason to take a critical look at things. Credible scientists (Phillip, Raphael, Julian etc.) are, very clearly, putting their reputations on the line. I would hope that most of us would agree that a quick particle prep is a small price to pay to resolve the issue.
LikeLike
@Nony, on the subject of “poor TEM”
Ong et al, ACS Nano 2013 “We believe that we image molecular features and not features related to the gold core, as high-resolution transmission electron microscopy images of these particles do not show any evidence of protrusions with these spacings (fig S12)”.
Fig S12 is not “poor TEM”: it is high quality EM showing a nice faceted nanoparticle.
Jackson et al, Nat Mater 2004, “Additional confirmation of the presence of ordered phase-separated domains was provided by transmission electron microscopy (TEM) images. In fact, in these images (see Supplementary Information, Fig. S2) we have found that there is an observable ring around the nanoparticles’ metallic cores consisting of discrete dots spaced ∼0.5–0.6 nm.”
In that case, it is indeed arguably poorer EM. David A Muller (Cornell) commented that: “Funny thing is the TEM image from their appendix, Fig S2a, that is cited as independent confirmation is also an instrumental artifact. etc”, see:
It matters little, whether the EM shows features from the stripes (Jackson 2004) or does not show any (Ong 2013), the conclusion is the same: it’s still a duck: http://imgur.com/gallery/1BXxi
LikeLike
Nanonymous,
Indeed, the 2004 images are generally valid (despite that there may have been issues with feedback-loop artifacts for at least one of them) and have been reproduced. And the arguments of Moriarty and co-authors do fall apart, as I have shown since my first post on PubPeer.
The stripe features can be distinguished from noise, as the Radon images and other analyses demonstrate. It is not a subjective matter. The noise can be high, but the stripe features can (and have repeatedly been) distinguished from noise.
I don’t think that if Moriarty imaged a batch of particles the issue would be settled. As it has become evident on this blog, Moriarty, Levy and Stirling have again and again handpicked arguments in an inconsistent manner to justify their preconceived hypothesis. They are not only deceiving the non-experts, but also continuously deluding themselves. Fortunately, peer review will in the long run put Moriarty and Levy in their right place: as persistent fanatics not unlike those behind conspiracy theories. Unfortunately, they make a lot of noise and capture an inexperienced following as they are pushed toward the fringes of the field.
Clearly, who in his or her right mind would send samples to people who are ready to misinterpret the findings so as to validate a preconceived position? Samples have been sent to credible labs: Renner and De Feyter have also put their reputations on the line. Instead of partnering with credible STM scientists as Stellacci has done, Moriarty and Levy side with a graduate student who has not obtained a PhD and has not a single relevant publication to show.
LikeLike
Your arguments fall apart pretty quickly once you realize that all images from 2004-2009 indicate strong feedback oscillations. Particularly, the tunneling current in images from 2004 oscillate 1-2 orders of magnitude from the set point. Stellacci did share his data, quickly withdrew it from his website — a not so professional move, at least not while the controversy hasn’t settled yet, after all, what is there to hide?— but you can find a backed up copy on Raphael’s blog. Instead of handwaving with your pseudo arguments and ad hominem attacks, I strongly advise you to take a look at the files yourself. Apparently, you haven’t done so.
I would like to re-emphasize that I have strongly criticized the work of Stellacci purely on the scientific basis. Back in 2005, I didn’t know Raphael. At the end of 2012, long after I had forgotten the whole ordeal, one MIT faculty member notified me that my concerns from 2005 are now in the public domain. Only then, I contacted Raphael and asked if I could write a short guest post recounting my experience with the subject matter.
Finally, I’ve seen the same ad hominem attacks “…graduate student who has not obtained a PhD” repeating all over and over again. I think everybody would agree that obtaining a PhD is an admirable and certainly reputable achievement, but deciding to pursue carriers outside of academia — and not completing a PhD, at least not yet — is not indicative of inferiority and lack of credibility. In the matter of fact, Retraction Watch is full of falsified and misrepresented publications from renowned scientists from every possible discipline — some people simply choose to cheat to get by and advance their careers faster.
Attempting to assign lesser credibility (repeatedly) to a grad student who was at the epicenter of the controversy back in 2005 speaks volumes about strength of your arguments. You made the right decision to post anonymously.
LikeLike
@ Predrag
A minor point – you are right, Stellacci did take down the public data a while ago, but I just checked and its reappeared now (http://sunmil.epfl.ch/), it must have gone back up sometime after Levy started mirroring it.
LikeLike
@ Predrag: excellent points. I am truly honored to have you as a contributor to this blog and as a co-author on the paper.
LikeLike
@ AC check again: the links have always been there but they don’t work (error 404)
LikeLike
@ Levy & @ Predrag
You are right about the raw data being gone at Stellacci’s site, I just saw the links and assumed they were still up. I apologize completely – Predrag is right, not so professional, almost as though they don’t want everyone looking through it!
LikeLike
Djuranovic,
Your points are not supported by the data in the paper you have co-authored with Stirling et al. You can’t claim that all of the images from 2004 to 2009 suffer from feedback loop artifacts; first, it is simply inconsistent with what your paper with Stirling claims; second, there are probably hundreds of images behind the data published in 2004-2009, and in the paper you have only analyzed ONE for feedback-loop oscillations.
I agree with you in that not having a PhD or publications doesn’t ***by itself*** diminish anyone’s credibility in a scientific discussion (it would however matter in a grant or job application, where past performance matters). However, the fact that you don’t have a PhD degree and that you have not published a single paper after so many years of doing research ***while making basic scientific mistakes in a blog post and in a preprint with Stirling et al.*** DOES diminish your scientific credibility.
But let me elaborate on this. Having or not a doctorate, and having zero or many publications matters little when experts discuss on scientific grounds. What really matters in a technical discussion are arguments, not status. Actually, this is why I have not signed my posts, as I want my arguments to speak by themselves; putting the the weight of my position and carrier behind them could be seen as unfair to Stirling et al.
However, for non-experts participating in the discussion, such as Nanonymous and Nony, past achievements do matter because they do not have the necessary expertise to judge by themselves the validity of the technical comments. This is like when one goes to the doctor. You trust that the checks and balances of the healthcare system have only allowed people with the appropriate expertise to take care of your health. Surely, there’ll be cases whether the healthcare system has failed and lets an unprepared or irresponsible doctor practice medicine, but in general it works well. The same with the checks and balances of the academic system: grades, publications, peer review, and so on. For non-experts, lacking the ability to judge the technical expertise, they have to assess what the system has put in place (degrees, publications, grants, prices, etc.). in order to triage the good from the bad from the ugly. So when I mentioned to Nanonymous that Levy and Moriarty have partnered with you instead of with someone with proven credibility, that’s what I meant, that they have chosen the ugly.
Indeed, they have chosen you, someone who makes the blunder of arguing that because stripes arising from ringing look similar to the stripes published by Stellacci, then the stripes published by Stellacci come from ringing. Indeed, that’s all you argue in your post here: https://raphazlab.wordpress.com/2012/12/11/seven-years-of-imaging-artifacts/ What about tip speed independence? What about rotation independence? What about stripe spacing? These are the things that one should look at to distinguish features that are genuine from those arising from ringing. This is what Stellacci has analyzed in various papers. Instead, you just compared how images look.
LikeLike
@Unregistered:
I am going to refrain myself from discussing scientific credibility. The technical matter can be analyzed by undergraduates at MIT (or any other school). I can detect that you are trying to divert the discussion into this murky territory where the matter becomes less factual.
While in Stellacci lab, I’ve analyzed hundreds of images recorded by Alicia Jackson — published and unpublished. Unfortunately, absolutely all suffer from feedback control issues (oscillations). The analysis of feedback loop issues is covered at length, although erroneously, in the JACS paper. Stripies are not scan speed/angle independent, as claimed, and one wonders how such results were obtained. Our paper reanalyzed the same data and finds the contrary. And you could see it yourself, had that link on Stellacci website actually worked. Then, something changed in 2009. Feedback ringing becomes less of a pronounced feature in images (I’ll assume that Alicia Jackson graduated and that the new student finally corrected feedback parameters), so stripies had to be reinvented by image processing (offline zooms and misleading image manipulations and interpolations). And finally, in 2013, after proper image acquisition — ten years after the first stripy work took place — stripes completely disappear (to the human eye). And then comes the whole PSD analysis, covered at length in our paper.
I really can’t help you understand the issue better than to implore you to download free software Gwyddion and ask Stellacci to send you 2004 Nature Materials data. I’ve seen it. Only then you will discover the “ugly”.
LikeLike
Djuranovic,
You just claim things without providing support for them. For example,
– “While in Stellacci lab, I’ve analyzed hundreds of images recorded by Alicia Jackson — published and unpublished. Unfortunately, absolutely all suffer from feedback control issues (oscillations).”
Can you prove this? In your paper you only show potential ringing for ONE image.
– “The analysis of feedback loop issues is covered at length, although erroneously, in the JACS paper.”
Why is it erroneous? What is actually erroneous is the analysis in Figure 4 of your paper; the comparison of error ranges in your Figure 4 is misleading, as I have explained in detail in previous posts.
– “Stripies are not scan speed/angle independent, as claimed, and one wonders how such results were obtained. Our paper reanalyzed the same data and finds the contrary.”
Your Figure 4 is misleading, and the summation approach in Figure 5 represents a basic logical blunder, as I explained in detail on PubPeer. You will have to properly address my points in a scientific manner, instead of writing unsupported statements like this.
– “And finally, in 2013, after proper image acquisition — ten years after the first stripy work took place — stripes completely disappear (to the human eye).”
I guess you mean to your human eye. Have you looked at the Radon images? They clearly show aligned features. By the way, the PSD fits in your paper have not been done properly, as explained in detail on PubPeer.
These unsupported blanket statements that you make suggest how careless you are with scientific reasoning and practice. This is probably an indication of how ‘careful’ you were when doing STM. Perhaps getting images without feedback loop ringing was beyond your ability?
LikeLike
I am left utterly puzzled by your perspective, you keep iterating that the images have been reproduced and mere inspection of the set of figures in this post demonstrates that they have not.
The tone of your writing is suggestive (“fanatics”, “fringes”, the (ridiculous, think about it) suggestion that a graduate student is not credible because he hasn’t published a paper). I can’t comprehend why you think that sending particles would be a bad idea, I think the magnanimous response would be to send the particles with some images and careful explanations in a pleasant tone. If their interpretations are so crank-ish, why not encourage themselves to hang themselves with their own rope? Readers wouldn’t have to listen to their interpretations of the images, they could make their own judgements with the images side by side.
In any case, the discussion now borders on absurdity between us. Why would a graduate student who hasn’t published anything not be credible? I can find you many students of immense talent who have not published. There are many more factors in a graduate student publishing a paper, I don’t consider not publishing any sort of fault.
I doubt this discussion can go much further. I cannot reconcile your statement that the 2004 images are valid and that they are reproduced: from the post, images 1, 2 and 11 are *categorically different* than image 4. If they are images of the same sort of object, than something is very very wrong with the way the images have been acquired. This conclusion requires essentially no scientific training, there is no subtle piece of scientific knowledge that would help one reconcile the dissimilarity. I feel like I am returning a parrot.
LikeLike
Nanonymous,
The 2004 images have been reproduced by Stellacci in many publications (see JACS 2006, for example), and Renner and de Feyter have as well (see the comparison of images taken at different labs in Fig. 1 of Biscarini et al. and the PSDs in the same paper).
To nonexperts like you, just looking at images side by side without knowing much about the conditions under which have been acquired can lead you to the wrong conclusions (and Levy and Moriarty use this to their advantage). The 2013 images, such as image 4 above in this thread, are not meant to *look* exactly as the 2004 images. The 2013 images are from high resolution STM and are meant to show single ligands (the bright blobs). Instead, the pre-2103 images were taken at low resolution and can’t resolve single ligands; instead, they show coarse-grained contours. What you should look in terms of what is common among these images is alignment (of countours and blobs) and geometry (organization and spacing of the blobs). It is obvious that there is feature alignment in both the 2004 and 2013 images, and Biscarini et al. have shown that the geometry also matches. Also, I have demonstrated with the Radon transform analysis that the alignment is there for all these images, and that it can be distinguished from noise.
LikeLike
Unreg:
I’m a non-expert in colloids or SPM. But I’m middle-aged, won a national award in material science as a grad student, and have published and worked in many fields of science and engineering. I know how to look at things, even outside my expertise, and even how to hold my own lack of expertise in the balance.
Your firing off about validating earlier work (when not clear comparables) and your ad hominems are very much more what I would expect out of politicized parts of science or business.
The stripes get fainter when the resolution is better (huh? and that validates the original…huh? and the systems are not even the same sometimes?)
Also the not sharing the particles, the HUGE fight to even share SOME data…is bad news. You can say it is ‘because those baddies just want to find problems’ or even ‘they won’t do proper analysis’, but withholding of data is BS. It’s like playing an online game with someone who stalls it out or cheats because you said something in chat (sometimes even you didn’t…these cheater/stallers just always play that game).
FS smells bad. Tunneling current 2 orders of magnitude jumping around? Sigh. He’s going down! 🙂
LikeLike
Unregistered,
In this comment you state that the only difference is the resolution of the scan, can you explain why we can’t we replicate the general look of the 2004 image from the 2013 image by mere subsampling/lowpass filter?
LikeLike
Nanonymous,
You can’t replicate the general look of the 2004 image from the 2013 images by image resampling because the 2013 images show ligand tail groups while the 2004 are rough contours. The precise topography that the tip ‘sees’ is different!
LikeLike
So WHERE are good images (not crappy tunneling current feedback bouncing around 100 times value) that DO duplicate the 2004 images?
The 2004 images are feedback crap. And the 2013 images don’t show stripes. You got nothing, dude! Nothing has been shown! Ball in your court…
LikeLike
Unreg, the fact that you can’t resample a high res image and not get a correct low res image is troubling. Even with cameras, I’m sure there are various small effects between cameras (color shifts, chromatic aberration etc.) so that downsampling a high res image from one camera won’t line up *exactly* with a low res image…but there is no reason to believe that the general value shapes and contrasts should be anything more than a monotonic transformation between the two…any gross feature in a photo that could be seen in low res and not explained in the high res image (or replicated in a downsampled version) would suggest that at least one camera isn’t working terribly well.
I’ve asked before but no answer, can you provide an analogy to another field of scientific imaging wrt to downsampling not working and not deriving conclusions from not reproducible features from image to image? I can’t think of any.
LikeLike
Ugh, retyping last para:
I’ve asked before but no answer, can you provide an analogy to another field of scientific imaging wrt to downsampling not replicating features in images that were acquired at lower res and deriving conclusions from features that are not reproducible from subsequent scans? I can’t think of any.
LikeLike
Nanonymous,
Once more, resampling will not get you anywhere because the 2004 and 2013 images are imaging different things! Coarse grained contours show up as ripples; ligand tail groups appear as blobs. You have to understand that STM images are not typical photos where you get what you ‘see’. In STM you have a tip that is tracing the sample line by line. Depending on how the tracing is done and the trajectory of the tip, the tip will ‘see’ different things. The countours of a row of ligands and the tails of these ligands are two different yet related things.
LikeLike
UnReg,
I guess your point is that “resolution” in STM doesn’t mean what resolution typically means with other imaging methods….I don’t know enough about STM to say much to that, it certainly sounds odd to me though. Can you provide an example unrelated to Stellacci’s work where one can clearly see a “zoom in” to see this effect (coarse features that disappear at high resolution because the STM is “seeing” something different)?
Can you provide an analogy for the other point you have used in this discussion, that is, not being able to image the same feature on subsequent scans in the same location does not invalidate the image feature? Especially since we don’t seem to have a particularly detailed physical model as to how the feature is moving. Most scientific imaging methods rely on signal averaging to remove noise, a cursory glance some STM introductions and textbooks suggests the same for STM. At one point you used an example of dice, which seemed rather contrived. Can you cite analogous example (unrelated to Stellacci) in STM and ideally another imaging example outside of STM? I presume there would be a simple analogy that would make sense in, say, astrophotography but I can’t think of one .
LikeLike
I just wanted to add a note following the release of the Science article.
Unreg,
You’ve consistently projected Stirling et al.’s position as clearly wrong and uninformed. You state that there is nothing to reconcile between the 2004 and 2013 images (in spite of seeing a material acquisition error in one image). You’ve clearly stated that the group’s analysis is suffering from elementary errors and blunders and that the group has deliberately engaged in obfuscation.
The science article quotes another STM expert not connected to the group clearly:
“[Kevin] Kelly agrees with critics that the stripes in the original paper “look like an imaging artifact,” but he and others say the jury is still out on more recent reports.”
It appears that there are other experts in STM who seem to be suffering from the same logical blunders and errors in thinking.
Either you do not have the expertise you imply or you have not been honest about how an STM expert might interpret the 2004 images. The validity of the 2004 images is clearly questionable (apparently, by mere inspection from an expert), and your arguments against that position do not seem at all credible. I find it hard to believe that you much expertise in this field, given that you never articulated your position in the form of “well yes, the 2004 images look a lot like artifacts and you wouldn’t be wrong to worry about that, but here are some very good reasons why I know they are real…even an STM expert might miss them”.
LikeLike
I agree with nanonymous.
LikeLike
Unreg:
Please stop! You are just making a fool of yourself repeating the same false things again and again. “you can’t see the stripes anymore if the resolution is improved” that’s one of the most ridiculous justifications for non-reproducability I have ever heard (and I have heard people blaming the moon…). Moriarty, Levy and Co have done an excellent job trying to correct a mistake in the scientific literature and they have tried to do this in a more modern way (mostly through blogs and online discussions) instead of the old-fashioned one (also because they were not allowed to do that by editors). I always thought online forums/blogs etc. could be the future for scientific discussions but after following the thread here and over at PubPeer I don’t believe that anymore and this is because people like Unreg can – with a minimal effort – make it impossible for a non-expert to decide which side is more convincing. I find this very sad because this story is very important since it highlights unfortunately many things that are broken in the scientific/academic ‘system’.
I have acquired 1000s of SPM images over the years and I am 100% sure Moriarty & Co are right. End of story Unreg!
LikeLike
Nony, formerSTMguy,
You have nothing substantial of scientific nature to say. Your words clearly denote that you are faithful believers, and as such contribute little to the discussion.
LikeLike
I’ve made comments on the specifics. (Yeah, a little levity, but so what…I gave you street cred for the Radon work…and you’re not perfect with the going after Predrag on the Ph.D…I mean should I go after FS for not getting tenure?)
FWIW, I just went back and reread the Arxiv paper. (I find rereading things, I understand them better later.) Some things that stood out on the reread:
-stripes extending between the particles, but hidden (intentionally or not) by too high off a contrast blacking out that part.
-low number of overall images shown and how selecting certain ones from a set (by FS) could bias the result
-change in resolution over time (even though good STM was very possible in 2004).
-the ripples on the stripes (not just stripes, but the sort of chevron character of them (looks non physical, and no explanation other than artifact)
-the very small size of the peaks in the Fuorier analysis (just shoulders really)
-How the stripes got LESS distinct (becoming unseeable in some of those smooth eggs) with better resolution. This is unlike my experience with increasing magnification/using better SEMs when looking at carbon black composite materials. (Then I see better, more features, etc. when I have a more powerful microscopy)
-The preponderance of criticisms
-The thoughtful explanation of STM confounders (and how it was not done in the FS Nature article)
-FS sharing the data and then unsharing it (and not sharing it all) and only sharing some grudgingly
-promising to share the particles and then not sharing them (that’s BS to say…they’re mean to me…he is HIDING from examination by CRITICS).
P.s. You probably believed Lance Armstrong too…not me. I threw the BS flag years before it was cool.
LikeLike
Nony,
your latest comments clearly show that you don’t understand the technical points; you don’t even bothered to phrase the arguments that you copied from the paper in a proper way. Again, you are adding little value to the discussion.
LikeLike
unreg: yeah maybe. But I’ve been around long enough to read things and evaluate them. you are overcertain on the faintest of faint stripes and have NO EXPLANATION for why the 2004 pics are not duplicated. Show me, show me.
Oh….and your ‘totally repudiated’ and the like sounds like Mike Mann.
LikeLike
Nony.
Your summary of the key arguments in our paper is spot on. You clearly put across the technical points in a concise and very helpful fashion.
Thank you.
Philip
LikeLike
thanks man. I was actually worried that I was hurting you more than helping you. with my BBS antics. And not acting all buttoned up and all.
Still…if we are ever in town, I will buy you beers and tell you cool sea stories and you can do the like.
Still remember being a chem gs and taking some physics classes…and they all thought they were better than me. And I kinda played into that shit a bit. And then when we had the first test, I got a 90+ and the next up was in the 60s. Buncha math snobs. Think I can’t handle the Bessel functions? Heck, I learned them in the Navy. Jo and Yo. Look em up in tables. Not even anything to do with science. Just operating the reactor.
[Edited by Raphael 26/01, 22:45 – sorry Nony, I can’t accept that last paragraph]
LikeLike
Nanonymous,
I already mentioned a few times over at PubPeer that Figure 1a in the 2004 paper indeed looks like it could have come from ringing. But I have also said repeatedly that this is only ONE image of the tens of images behind the 2004 results in the paper (including Figure 6d). The 2004 TEM work was done with nanoparticles of different substrates, with different operators, varying tip speed and different tips.
As your comments clearly demonstrate, you are not an expert in the field. Your statements of support of Levy and Moriarty are leaning towards what one would call unconditional trust. If you can’t assess the technical points properly, you should refrain from claiming that Stirling et al. are right. Or if you do, you should acknowledge that this is based on trust or faith, but not factual evidence, which without doubt you can’t properly judge. In this case, you are then joining the likes of Nony (who as is obvious from the comments above can’t even articulate a coherent story) formerSTMguy (who is a 100% believer according to his words) and Djuranovic (whose track record and technical knowledge have severe limitations, as his comments here and on PubPeer reveal).
LikeLike
tens of published (??) non ringing (but showing clear, strong stripe) images?
LikeLike
@Unreg:
the fact that you haven’t analyzed the images that Stellacci shared on his website (and are now mirrored on this blog), you have proven your “technical knowledge limitations”. Scan speed independence from JACS 2006 paper can not be reproduced, even when images are erroneously extrapolated to the higher pixel density (please, demonstrate the opposite with data). When you look at the tunneling current signal, is oscillates within 1000% of the tunneling current set point (for this particular set of images, in other pre-2009 images it’s even worse). Therefore, this data can be freely discarded, as these are not images of nano particle ligands ( perhaps not even nano particles: the images of plain gold foil are remarkably the same — visually and numerically, as calculated by surface roughness).
So far, you have only discredited yourself with non-scientific accusations and pseudo-scientific arguments. Perhaps that’s why you are posting anonymously and hiding your credentials. Yet you find it easy to speak of other contributors’ credentials.
To recap, you comments show the lack of understanding of basic SPM and your credentials remain unknown. If I were to judge you by your comments, you are either completely biased or complete naive to the basic concepts of experimental physics (data acquisition and analysis).
LikeLike
Djuranovic,
[moderated, see above]
LikeLike
Dear NonReg, I am not lying! I did the analysis myself in two different ways: with ruler (that’s how Stellacci and co-workers did it) and FFT. Spacings —on “nano particles” and “gold foil”, as the two are indistinguishable — in the image are invariably scaling with the scanning speed.
I am sure you have more imaging experience than me — that’s not hard given my short exposure to the subject matter — but your conceptual defficincies are rather staggering.
Shall we exchange our own reproductions of Figure 3 JACS 2006 to gain some insight into discrepancies?
But I am glad we’ve come to agree on one point: imaging of molecular features on curved surfaces is difficult with STM/SPM, most likely due to feedback issues. Lowering the scanning speed and reducing the scan size would tremendously help the stability of imaging. However it is rather unfortunate that Stellacci made beginner’s SPM imaging mistakes in initial papers and now has to go at length with all sorts of auxiliary analysis to attempt to confirm the hypothesis that may not even be true.
LikeLike
Unreg,
You do not address my requests for an analogies in STM and other fields of imaging and are not addressing the content of the quote. Another unaffiliated expert in STM agrees that the original evidence for stripes is probably an artifact. How much clearer can one make it?
I *never* claimed to be an expert in this field (I’ve stated numerous times that I know very little about STM); what in the world are you talking about in your post? I don’t know why you keep parroting your statement about “ONE image”, the expert opinion of Kevin Kelly suggests that the STM evidence in the 2004 paper is mere artifact, this is a clear statement about the validity of that paper (not just “ONE image”). It is isn’t difficult to follow the remainder of Stirling et al.’s chain of reasoning.
The trust is not at all unconditional, I’ve looked at their arguments critically and have seen expert testimony (IN THE QUOTE YOU HAVE NOT ADDRESSED).
I can see you are not participating in this conversation sincerely, I don’t think anyone can say that I didn’t try.
LikeLike
Nanonymous,
Kevin Kelly’s quote only says that Kevin thinks that the 2004 stripes LOOK LIKE an imaging artifact. I have also said so. They indeed “look like” artifacts, but Stirling et al. have only looked at the current signal for ONE image. But Stellacci, Renner and de Feyter have shown in 2013 that the features of 2004 are reproducible and that the PSD analyses of the 2004 and 2013 match. This is strong evidence that even if the 2004 images “look like” arising from ringing, they are not (although Figure 1a in particular in the 2004 paper may indeed suffer from ringing; but the quantitative analyses in the paper are based on tens of images of nanoparticles on different substrates, with different operators, varying tip speed and different tips).
As for the analogies, you can look at AFM, KPFM and STM papers of single-molecule features on curved surfaces. You will see similar difficulties in reproducing the features.
Because you are not an expert, [moderated, see comment above]
LikeLike
Unreg, you question the technical knowledge of others but say: “The 2004 TEM work was done with nanoparticles of different substrates, with different operators, varying tip speed and different tips.” So TEM is done with a tip?? And that tip is scanned at varying speed across the sample?? How dare you question the technical knowledge of others when you obviously do not have the background to understand these things. This is exactly what I meant when I wrote above that PubPeer is not the future of scientific discussions because people like you – who very obviously have no clue what they are writing about – can make clog the site with repeated (and false) comments.
I never wrote that I am a ‘believer’ of Moriarty & Co! I wrote that ‘I am sure that they are right’. This is completely different, because it means that – based on my own knowledge in the field – I come to the same conclusions as they do. The only difference is, that I didn’t invest all that effort (but I am glad they did). Another example of your approach to twist the facts and render the discussion impossible.
LikeLike
In his(her) defense, he probably meant STM, instead of TEM. If not — and I can’t imagine this scenario — we’ve been all waisting our time.
LikeLike
formerSTMguy,
[moderated, see above]
LikeLike
Unreg aka troll
I don’t like arguments of authority (such as the ones you repeatedly make re who has or has not a PhD, who is an expert or not, etc), but I will make an exception. This is my blog and I have had enough of your trolling. If you continue to just repeat the same things and make ad hominem attacks, I will not accept your contributions anymore.You are welcome to set up your own website detailing why you think our arguments are misguided and rebutting our paper and the comments here. I even promise that I will provide a link from the “stripy outside” page.
I give you a last chance though. You mention that although Fig 1 of article 1 is indeed a ringing artefact, there are other images, in that paper (‘behind that 2004 paper’ ??) which show that thankfully the particles in reality look like the artefact. Please give us the raw data for what you consider to be a convincing image from that data set. Thanks.
LikeLike
Levy,
[moderated, see comment above]
LikeLike
I’ve been following this debate for some time, and followed the data and analysis, and come to a clear conclusion. I’ve talked to lots of colleagues, who broadly agree with me. In fact, I’ve only met one who doesn’t agree with me, and he has links to the Stellacci work. I have discussed this with nanoparticle experts and one notable guy said everyone knew this, it was just a matter of time before the opinion came out. A little about me – I’m a nanoparticle research professional – I have an international profile for my work and although I’m not the biggest guy out there, I know my stuff, and my words carry weight. I do not know any researchers on either side of the argument, but know their work well. When I first saw the stripy work, I was surprised, but not being a STM guy, I thought there must be a new technique I didn’t know about. Why am I anonymous? Because this isn’t my fight, and I don’t want to get overly involved. I’m going for tenure, and can’t be seen to be choosing arguments.
The facts are these – the analysis now goes well beyond what Stellacci originally published, which is clearly wrong. This is now sliding into a desperate fight to save a career that was unintentionally build on wrong results, and it’s getting personal.
This is now a case of what is well known as ‘pathological science’ -desperately clinging onto wrong results despite the overwhelming evidence to the contrary. The only reason this has not been retracted is because the work has been cited thousands of times, it will ruin Stellacci, and become a very public downfall, like the Schon case (with the exception that this is not intentional fraud) and no one wants to see that.
LikeLike
score board
LikeLike
It is good to hear an opinion that is essentially supporting the case of Stirling et al.
I think a discussion of word “intentional” in these situations deservers some discussion. We have evidence of testimony from an ex graduate of Stellacci that says that credible evidence against the stripes was demonstrated years ago. Predrag has (understandably) not gone into details regarding his interactions with Stellacci and focused on scientific criticism, but many of us who have been to graduate school can confidently guess the general nature of the events that transpired (given the outcome). Levy has been voicing concerns about this work for years. The journal’s long response time is also concerning (as is the appearance of an editor on this blog).
Clearly, it is difficult to say that whatever has happened hasn’t involved “intent” of some sort.
I’ve been saying this for awhile, but there are just so many similarities between this saga and arseniclife…refusal to engage in very efficient public discussion, adamant denial that doesn’t address facts and significant evidence of a consensus among peers that there is something very very wrong (although not quite as obvious here, few people seem to be willing to put their names (like you and I*) to these opinions…very hard to find people supporting the stripy work that are not directly connected to it). NASA and the journal Science have done a great disservice to the entire world in the case of areseniclife, both organizations have deliberately avoided any corrective action in the face of what can only be described as incredible stupidity. It is easy to become cynical of what we can expect from such organizations in terms of telling the plain truth and one gains very little by critiquing them (only really safe if you already have your Nobel prize).
Stellacci’s references to bullying and suicide are not what I would expect from a professional who was dealing with (what he presumably sees as) cranks attacking his work. These cues are enough for me to guess what has happened.
Understanding the network of social cues behind publishing scientific fiascos would be very interesting. I suspect they would have far more predictive power than trying to understand the actual work from a detailed technical perspective.
*in truth, there is no internet anonymity. Any reasonably aware person is now certain that one must assume that anything they post on the internet can be attributed to them; and there is little doubt that the privilege to access that information won’t be abused.
LikeLike
I’m not sure the right place to put this comment:
Reading through the 2004 paper right now (yeah, I know…mea culpa ;-)). It really comes across as what I stereotype as Nature-Science hype paper. Really it does. The abstract that reads more like an intro para (or a press release). “Unprecedented”, ‘we do both’, etc. Just the whole tone of a typical gushy Nature paper. Easy for me to say (as not pursuing a tenure track career), but I think typical person better off publishing in an ACS journal (the ones below JACS are quite good) or Physical Review (if you want prestige AND rigor). You can show all the data and also, you don’t have to write the thing as TEH COOLEST THING EVAH!
Also a lot of comments about having tested for artifacts and the like, but then not showing the data even in supplemental materials. Furthermore, there is no follow-up journal publication with full details (in a non space limited journal, say Chemistry of Materials). We do have years of different publications, but they really just have strands and snippets of info and don’t fully back up the Nature paper, like a normal full original study. It’s more of a bunch of straw trying to make a brick and not much solid earth. Extending the work, while trying to buttress criticism.
I actually suspect that some of the strong words about image artifact testing indicate that probably the referees DID question the work and that FS added that stuff to respond. And then the editor just threw it over the bar into the magazine. Well, now it gets ripped to shreds. And really, the thing to appease the referees would be a proper paper that showed the DATA! (this is a classic failing of NPG/Science papers…crappy methods sections and claims not backed up by data).
I guess it could be OK (it’s sort of the norm for Nature-Science) IF IT WERE TRUE. But if wrong, it really implicates NPG and just shows more of the problems we have seen with the glossies (and add into that, not publishing the criticism paper, not pushing for all the data, the [initial] excusal of the image re-use, the image reuse itself, Pep’s behavior, etc. At the end of the day, when the stripey is revealed to be a naked emperor, where does that leave NPG that they published it and that they did not support/allow critique (even fought it)?
I also see more the pattern of the nano-hype field (sorry Rapha) that I am used to from late 90s (when FS was being trained, btw). I just know that field from the late 90s. Everything’s being “tuned”, everything’s a sensor…it slices, it dices…it’s a Ginsu knife! The magic world of chemists claiming to make devices. I don’t see the same level of silliness in normal experimental fields like solid state chemistry, organic synthesis, etc
LikeLike
Perhaps “Unreg” should comment on the re-use of data across a number of papers from the Stellacci group, before attempting to extract patterns from noise? If s/he requires a refresher course, they might want to read my most recent post on the subject
and then frame a suitable response.
LikeLike
(Sorry for the stream of consciousness posting.)
Figure one of Jackson has a very long caption. One part says “Particular care was used in assuring that the observed nan ostructures were not imaging artefacts…Second, during each imaging session, images were collected at scan rates and scan sizes that covered a range of values from 3–6 Hz and 60–200 nm, respectively. Periodically, the scan direction was varied.”
Now, when I read that it sounds like they really did do some good checks of the images. I wish they SHOWED this info…but at least an assertion of controls and care is made. But I get the impression (not sure if I’m following) that they didn’t actually re-image the same places (!) to show features invariant? If so, this could be rather dishonest. I wonder if the text was added to appease critical reviewer. But it gives an incorrect impression.
LikeLike
Nony, good observation, the data isn’t showed, rather it is verbalized to appease the reviewer ( thats my guess). From the Stellacci data release, we know that “stripy” features are NOT scan speed independent, despite their assertion that they are. Anybody can check that — It’s now public. It’s the house of cards thereon ….
LikeLike
1. I would be interested to know which Nature editor handled the Jackson paper. Pep?
2. The article says “Received 9 December 2003; accepted 15 March 2004; published 18 April 2004”. Does this mean it was accepted without revision?
LikeLike
Hmm…kudos to the FS team if they got that thing in without revision. But…I wonder. Wonder if there were some edits done (not shown) during the NM20004 editorial period. Or perhaps that paper had earlier been submitted to a different journal (Nature overall? Science?) and then changes made based on the review cycle there.
LikeLike
Hi, Nony.
As Pedja says, the claims don’t stand up to scrutiny. “Particular care” was certainly not taken. At no time for the Jackson et al. 2004 paper did Stellacci and co-workers attempt to image the same particle twice. The statement in the paper is highly misleading.
A representative set of the data collected by Stellacci and co-workers for the Nature Materials 2004, and subsequent papers, is shown here: http://www.nottingham.ac.uk/~ppzpjm/From_Striped_Nanoparticle_Archive.pdf
No credible scanning probe microscopist would draw conclusions regarding the spacing of molecular features on the basis of this data.
Philip
LikeLike
They look really bad, Phil.
LikeLike
Interesting that the Biscarini, et al Langmuir 2013 paper was reviewed in two weeks. Seems awfully fast unless at a conference. (Not making some huge inference, just something I noticed and found mildly peculiar.)
LikeLike
Julian, feedback on the PSD pdf:
Very nice and wow…that whole curve fitting looks very sketchy to extract info from. They really don’t have peaks, they have shoulders. And other than the things going generally downwards, I struggle to say the curves are similar (the undulations that is). Would Physical Review put up with this stuff? Seems very handwavey just even trying to read into these little wiggles. And then what you show on the fitting and the black curve and just the ones that don’t look similar but are claimed similar. Just seems like bad news.
I would urge you to hone that presentation AND THE PAPER ITSELF to be as explanatory as possible. Remember this stuff is complicated and hard to follow. The situation here is different than a report of non-controversial findings in a paper journal, where a very clipped format is used. Very important to explain things here because that is the whole point of the paper.
Some tutorial things:
1. First set of images a-h along with the explanation with parens and such was very hard to follow. Consider to separate the ball windows from the graph windows and number differently. Also, it was confusing to have parenthesized statements when you are also referring to (c) and such. Also, the order of explaining a then diverting to c, etc. was confusing. Instead, why not show and explain the simulated mages first. Could caption them Sim-1 through Sim-8. Then show the result (Res-stripes, Res-dots) and discuss it.
2. The very first image you show from Biscarini needs more explanation. I was baffled. (rest of the explication went pretty well).
Science things:
1. I wonder if it is a fair test to show the striped balls only lying on their sides. Yeah, this is what the 2004 image shows, but that’s what made it unrealistic (no archery targets). Maybe show the PSD for random stripes on side and then if the balls have no preferred alignment as they precipitate.
2. Since the 2004 images had feedback ringing and visible stripes (we know that), it doesn’t surprise that they show a PSD feature. I mean, we can see stripes anyway.
3. for the curves, do they use overall images or cherrypicked sections. Also, do they use the raw data or offline-zoomed (therefore filtered and smoothed) data?
LikeLike
Hi Nony,
Thanks for the comments. I agree we do need to improve the clarity of a couple of the sections in the paper when it is revised. The paper was written after we had been absorbed in the work for months, now the argument have had some time to breathe in public I think we have a better idea of where the explanation needs improving. I also agree with both your points on reordering sub-figures and use of parentheses (I tend to have side thoughts and use lots of brackets [and then use nested brackets {until it all goes crazy!}]).
As for the science:
1. It is an interesting question, but making the 3D model in MATLAB, applying an arbitrary 3D rotation, and then calculating the surface topography would be very difficult. I would invest the time doing this if Stellacci regularly argued he could see these poles in his STM data. However, he does not, and I have lots of other work to do.
2. Yes we expected features in the PSD as we can see something in the images. But their argument is that the features have the same width even with different scan conditions, and that if the widths are unchanging then the features are real. This is why they fit the PSDs to try to show the shoulder positions are always the same. This is why the issue of biasing the fits becomes important. Also we should remember that between images that multiple scan parameters are changing (not just speeds), and that the images are not of the same area, so these are not fair tests.
3. My understanding of the paper is that they are telling us that this is the PSD of the full raw image. I do not have the raw data for all of the images in Biscarini et al. to check this. The data Biscarini provided for me was the PSDs and fits (the fitted curve, not the fit parameters) for the 2004 data.
LikeLike
I hope you can think of some ways to publish LPUs or connect this to your own program so that the investment is more than just criticism (to motivate you). Actually, I think digging into this stuff can have benefits over just finding some other system to image.
1. Come on. Smart guy like you? Maybe pull in some MATLAB-geometry jock. I donno…you have enough to go on to just show different microstructures can cause shoulders (would think even speckles might do this). But still would be cool to really start exploring that parameter space (have a distribution of ball sizes and geometries, etc.) I guess it turns into a whole investigation of PSD versus geometry, though.
2. I didn’t get that. Please explain it clearer. (maybe label their figure) 🙂
3. I thought Phil had the raw data? There may be non-optimal choices in the conversion of 2-D SPM image data to PSD (cropping, etc.).
LikeLike
(I realize it is easy for me to just say things on the Internet…that require others to do work.)
1. Maybe it would be interesting to have some of the 2004 raw data shown in the PLOS paper. Is it normal to have it so streaky and all or is that indicative of overall bad work?
2. Also, I think a graphic (or some math) showing the feedback current going negative and shooting up orders of magnitude. Perhaps show a comparable feedback excursion plot from other data (well regarded study of approx. 2004). I am imagining one that would be spiky and the other pretty flat. For graphical purposes, could use a semilog scale and show negative excursions as red.
LikeLike
I´m not completely familiar with this stripy nanoparticle debate (with AuNP, SAMs and so on though). What puzzles me is: if this stripes COULD be artefacts of STM measurements (nobody doubts that I guess?), how could STM measurements be acceptable as a proof for stripes in the first place?
And how could such “proof” pass the rigorous (?) peer review process of so many high impact journals?
LikeLiked by 1 person
“And how could such “proof” pass the rigorous (?) peer review process of so many high impact journals?”
Indeed. That’s a question quite a few of us have asked. See http://physicsfocus.org/philip-moriarty-peer-review-cyber-bullies/ and the links in that post for more on that question.
Raphael’s comment here also provides a relatively recent appraisal of the debate: https://pubpeer.com/publications/58199FBEA31FB5755C750144088886
LikeLike